ISSN 2410-5708 / e-ISSN 2313-7215
Year 12 | No. 35 | October 2023 - January 2024
© Copyright (2023). National Autonomous University of Nicaragua, Managua.
This document is under a Creative Commons
Attribution-NonCommercial-NoDerivs 4.0 International licence.
Genotyping of extended-spectrum beta-Lactamase genotyping in Salmonella spp isolated from ground beef in supermarkets in Managua, Nicaragua 2020
https://doi.org/10.5377/rtu.v12i35.16960
Submitted on 30 th August, 2022 / Accepted on 21 th September, 2023
Francisco Romero Oviedo
National Autonomous University of Nicaragua (UNAN-MANAGUA), Associate
Professor Clinical Bioanalysis POLISAL, Laboratory of Molecular Biology "MA.
Elmer Cisneros in memoriam" Managua, Nicaragua.
Oscar Arbizú Medina
National Autonomous University of Nicaragua ( (UNAN-MANAGUA), Associate
Professor Clinical Bioanalysis POLISAL, Laboratory of Molecular Biology "MA.
Elmer Cisneros in memoriam" Managua, Nicaragua.
Benita Magaly Jimenes
National Diagnostic and Reference Center
(CNDR), Nicaragua.
Yosimar Narváez Gonzalez
National Autonomous University of Nicaragua (UNAN-MANAGUA), Laboratory
Analyst Clinical Bioanalysis POLISAL, Laboratory of Molecular Biology "MA.
Elmer Cisneros in memoriam" Managua, Nicaragua.
Section: Health and Social Services
Scientific research article
Keywords: Salmonella, beta-lactamases, Meat, Food, Health.
Abstract
Antimicrobial resistance of foodborne pathogens has become a major global concern for human and animal health, bacteria such as Salmonella are the main microorganisms related to food poisoning and can survive constant changes in environmental factors, therefore, it was decided to search through a descriptive study this pathogen in ground beef. A total of 100 randomly selected samples of ground beef were obtained from 10 supermarkets in Managua, of which only 4 samples isolated Salmonella spp showed phenotypic and genotypic beta-lactamases, conferring resistance to cephalosporins and limiting the use of fluoroquinolones due to an intermediate resistance to ciprofloxacin.
Introduction.
Foodborne bacteria that present resistance to antibiotics have become a problem for public health worldwide, there is a concern from the point of view of human health and animal health due to food safety and food security, as well as economic implications for the most vulnerable sectors, since infections with resistant bacteria require more expensive antibiotics to control the infection. The Food and Agriculture Organization of the United Nations (FAO) has shown its concern after evidencing that food of animal origin can serve as a route of exposure to bacteria with antibiotic resistance, it is important from the epidemiological point of view to evidence the presence of Salmonella spp pathogenic for humans with resistance profiles of clinical importance. (FAO, 2020).
The World Health Organization has shown its concern by reinforcing scientific knowledge through research processes and epidemiological surveillance of member countries, which has led to a global strategy called “One Health” to contain this increase in antibiotic-resistant bacteria. (WHO, 2016)
Salmonella is a gram-negative, facultative anaerobic bacillus belonging to the Enterobacteriaceae family. The genus Salmonella is composed of two taxonomic species, Salmonella bongori and Salmonella enterica, and all medically important Salmonella are part of the latter. Salmonella enterica is a diverse species of bacteria consisting of more than 2500 different serovars. The pathogen can be host-adapted, host-restricted, or generalist, depending on the wide range of hosts it can infect. The pathogen is ubiquitously present in the human food chain and is often associated with outbreaks of foodborne illness. Salmonella colonizes nearly all cold- and warm-blooded animals, in addition to its extra-animal environmental strongholds. Recent decades have witnessed the emergence of highly virulent, antibiotic-resistant Salmonella, leading to increased morbidity and mortality in humans. The emergence of several Salmonella serotypes resistant to multiple antibiotics in food animals underscores a significant food safety hazard. (Divek V. T. Nair, 2018. )
Antibiotic resistance in foodborne pathogens such as Salmonella is a major concern for public health safety. More attention is needed to focus on them in the animal feed supply. Salmonella is difficult to eliminate from its reservoir hosts, and food animals often serve as reservoirs for the pathogen. Nontyphoidal Salmonella causes the largest number of illnesses, hospitalizations, and deaths associated with foodborne illnesses. (Divek V. T. Nair, 2018. )
Method
A cross-sectional study was conducted to detect genes encoding β-lactamases (OXA, TEM, SHV, CTX-M) in Salmonella spp isolated from ground beef sold in supermarkets in Managua, Nicaragua 2020. The universe consisted of 46 supermarkets located throughout Managua, distributed in 5 chains. The sample consisted of 10 supermarkets, in each of which two batches of meat were taken (one batch of economic meat and one batch of super meat), from each of the batches 5 subunits were obtained, i.e. 50 subunits of economic meat and 50 subunits of super meat were studied, which is equivalent to 100 subunits of ground beef studied. It should be noted that for each of the subunits, 100 grams of ground beef were analyzed under normal conditions of sale to the public. The samples were then taken to the laboratory of the Diagnostic and Reference Center of the Ministry of Health, and the type of sampling used was stratified random probabilistic (of the 46 supermarkets, 10 were obtained randomly, representing 20%). To obtain the sample, the randomly selected supermarkets were visited in the company of the Food Regulation Directorate of the Ministry of Health, using data collection cards for the collection of data concerning the investigation, based on the information observed in the establishment and the product. The 100 grams of ground beef were obtained for each sub-sample under normal conditions of sale to the public, then transferred to the laboratory of the Diagnostic and Reference Center of the Ministry of Health, where the analytical areas, reagents, and materials were provided to carry out the processing of the samples and the isolation of Salmonella spp
Isolation of Salmonella spp in ground beef
The isolation procedure for Salmonella in ground beef was carried out following the recommendations of Chapter 5 of the Bacteriological Analytical Manual U.S. Food and Drug Administration, Brief description. 25 g of the sample was weighed in a Stomacher bag and 225 ml of APB (buffered peptonized water) was added, and incubated at 37° ± 1° C for 18 ± 2 hours. The bag was shaken with the pre-enrichment medium (APB) and 0.1 ml was transferred to a 10 ml Rappaport- Vassiliadis tube and incubated at 41.5 ± 1°C for 24 ± 3h. The Rappaport Vassiliadis tube was mixed and shaken and transferred with a 3 mm loop an inoculum to the selective enrichment media XLD (Xylose, Lysine, Deoxycholate), HE (Hektoen Enteric) and SB (Bismuth Sulfite) then incubated at 37° ± 1°C for 24 ± 3h. After the established time, these plates were examined for characteristic colonies of Salmonella spp, according to the medium used. Three typical colonies of Salmonella spp were selected from the Agar plates and inoculated in the biochemical tests TSI (Triple Sugar Iron Agar), LIA (Lysine Iron Agar), Urea Hydrolysis, MIO (Mobility, Indole and Ornithine), Mucate, Citrate, Malonate, Methyl Red. Once the biochemical test reactions were positive for Salmonella spp, the microorganism grown in the TSI was transferred to a TSA dish, incubated at 37°C for 24 hr and the oxidase test was performed. For serological confirmation, polyvalent somatic antisera O poly A-1 and Vi. (Wallace H. Andrews, 2019). The genetic materials of the isolated strains were sent to the Laboratory of Molecular Biology “MA. Elmer Cisneros in memoriam”, Instituto Politécnico de la Salud, Universidad Nacional Autónoma de Nicaragua, UNAN-Managua for molecular analysis.
Antimicrobial susceptibility testing
The Kirby Bauer or Disc Diffusion method was used (Rodriguez, 2000), brief description: the suspension was prepared with sterile saline solution (0.85%) with an optical density of 0.5 McFarland of the inoculum in test tubes, using a densitometer (Densichek). The sterile swab was dipped into the prepared suspension and some of the excess was removed from the inner walls of the tube. It was seeded by the tri-directional streaking method on Mueller-Hinton (MH) agar. Sensidiscs were strategically placed for the detection of resistance mechanisms. They were incubated at a temperature of 37°C for 24 hours. The sensidiscs used were AMP (ampicillin), SXT (sulfamethoxazole/trimethoprim), TET (tetracycline), CRO (ceftriaxone), CTX (cefotaxime), AMC (amoxicillin/clavulanic acid), CAZ (ceftazidime), CIP (ciprofloxacin), MER (meropenem), IPM (imipenem). The reading of the halos was performed with a caliper, according to CLSI (Clinical & Laboratory Standards Institute) 2020 standards. (Clinical and Laboratory Standards Institute, 2020) Klebsiella pneumoniae ATCC 700603 and Escherichia coli ATCC 25922 strains were used as quality control.
DNA extraction
Bacterial DNA was extracted by lysis from the culture on MacConkey agar incubated for 24 hours at 37°C, a pool of CFU was taken, inoculated into an Eppendorf tube containing 100 µl of nuclease-free water, placed in boiling water bath for 10 minutes, then centrifuged at 12000 rpm for 5 minutes and 80µl of the supernatant was extracted, the concentration of extracted DNA was determined in Nanodrop lite 2763 (Samuel Vilchez, 2009).
Detection of blaOXA, blaTEM, blaSHV, blaCTX-M genes
PCR (polymerase chain reaction) for the detection of genes coding for β-lactamases was performed using universal primers, following the procedure described by (Hong Fang, 2018. DOI 10.1128/JCM.01943-07). The PCR amplification reaction was performed in a final volume of 20 ul containing GoTaq® qPCR Promega 2X, 0.2 uM of each primer, and 2 ul of DNA from the strain under study.
PCR amplification conditions
Thermocycling conditions were as follows, initial denaturation for 15 minutes, followed by 30 cycles of 30 seconds at 94°C, 90 seconds at 62°C and 60 seconds at 72°C, ending with a final extension of 10 minutes at 72°C.
Electrophoresis
The amplified products were revealed by electrophoresis on a 2% agarose gel stained with GelRed in 1X TBE buffer. Electrophoresis was run at 120 volts for 60 minutes, and the DNA bands of the different genotypes were visualized in a camera with ultraviolet light and photographed. The weights of the bands were evaluated with the positive controls using strains of Klebsiella pneumoniae ATCC 700603 for the blaSHV gene, Escherichia coli for the blaOXA, blaTEM, blaCTX-M genes, and Escherichia coli ATCC 25922 negative control, not carrying the resistance genes.
Results
Of the 100 samples of ground beef analyzed, Salmonella spp were isolated. In 4 samples, the 4 Salmonella strains had a resistance profile to the β-lactam group, ceftazidime, cefotaxime, and ceftriaxone, and also showed resistance to ampicillin, nitrofurantoin, tetracycline. The strains were sensitive to trimethoprim/sulfamethoxazole, amoxicillin/clavulanic acid, meropenem, and imipenem. ciprofloxacin (fluoroquinolones) presented intermediate resistance. Of the 4 extended-spectrum beta-Lactamase (BLEE) genes searched by PCR (blaOXA, blaTEM, blaSHV, blaCTX-M) only the blaCTX-M gene was detected.
Discussion
Given the increase in antimicrobial resistance (AMR), seen as a threat to the health of humans, animals, and plants, and the need for the WHO “One Health” approach, the need to know the prevalence of pathogenic bacteria resistant to antibiotics present in food intended for human consumption arises. There is little scientific evidence that food can serve as a vehicle for the acquisition of antibiotic-resistant bacteria and may therefore become the cause of many infectious diseases. Salmonella spp is a human pathogenic bacterium transmitted by contaminated food causing foodborne outbreaks and food poisoning worldwide.
A study in Lithuania showed a prevalence of 1.5 % of Salmonella spp in ground meats however no Salmonella with resistance to beta-lactam group were isolated (Margarita Terentjeva, 2017). Salmonella is not part of the intestinal microbiota of humans and when it is identified in humans, we could affirm that it is of animal origin, but we do not know if the resistance genes originate in microorganisms of animal origin, so it is necessary to continue investigating this behavior to look for clonal correlation.
Salmonella strains isolated in ground meats sold in supermarkets represent an alert because they possess the blaCTX-M gene that encodes enzymes capable of hydrolyzing antibiotics of the beta-lactam group, mainly cephalosporins, Although they are sensitive to trimethoprim/sulfamethoxazole, an option for treating diarrhea in Nicaragua, these strains are close to being classified as priority 2 on the WHO list of pathogens due to their intermediate resistance to a fluoroquinolone.
It is of utmost importance to know the dimension of this problem, since this behavior limits therapeutic options and contaminated food would be contributing to the dispersion of resistance.
Ethical aspects
since this was a risk-free study in patients, informed consent was not required and the identity of the supermarkets was kept confidential.
Acknowledgments
Lizeth Palacios López, Flor de Liz Vega Mayorga, and Danna Pulido Largaespada for their important collaboration with the study in the collection and processing of samples.
Abbreviations:
DNA: Deoxyribonucleic acid.
AMC: Amoxicillin/Clavulanic acid
AMP: Ampicillin
APB: Peptonized buffered water
BLEE: Beta-Lactamase Extended Spectrum
CAZ: Ceftazidime
CIP: Ciprofloxacin
CNDR: National Center for Diagnosis and Reference, Ministry of Health, Nicaragua
CRO: Ceftriaxone
CTX: Cefotaxime
FAO: Food and Agriculture Organization of the United Nations
HE: Hektoen Enteric Agar
IPM: Imipenem
LIA: Lysine Iron Agar
MER: Meropenem
MH: Mueller-Hinton Agar
MIO: Mobility, Indole and Ornithine Medium
WHO: World Health Organization
PCR: Polymerase Chain Reaction
POLISAL: Polytechnic Institute of Health
SB: Bismuth Sulfite Agar
SXT: Sulfamethoxazole/Trimethoprim
TET: Tetracycline
TSA: Trypticase Soy Agar
TSI: Triple Sugar Iron Agar
CFU: Colony Forming Units
UNAN: National Autonomous University of Nicaragua
XLD: Xylose, Lysine, Deoxycholate Agar
Funding
This study was carried out with the support of the Centro Nacional de Diagnóstico y Referencia del Ministerio de Salud de Nicaragua (National Diagnostic and Reference Center of the Ministry of Health of Nicaragua). Own funds Laboratory of Molecular Biology ¨Elmer Cisneros¨ in memoriam POLISAL UNAN Managua.
Conflict of interest.
The authors declare that they have no conflicts of interest.
Contribution of each author
Francisco Romero Oviedo A, B, C, D, F, G.
Oscar Arbizú Medina A, B, C, D, F, G.
Benita Magaly Jimenes A, B, C, D, F, G.
Yosimar Narváez González C, D, D, F, G
A-planning, B-information collection, C-information analysis, D-drafting, F-critical review, G-approval of the final version.
Objective
To identify genes coding for β-lactamases (blaOXA, blaTEM, blaSHV, blaCTX-M) in Salmonella spp isolated from ground beef sold in supermarkets in Managua, Nicaragua 2020.
Anexos
Figure 1.
1.5% agarose gel electrophoresis.
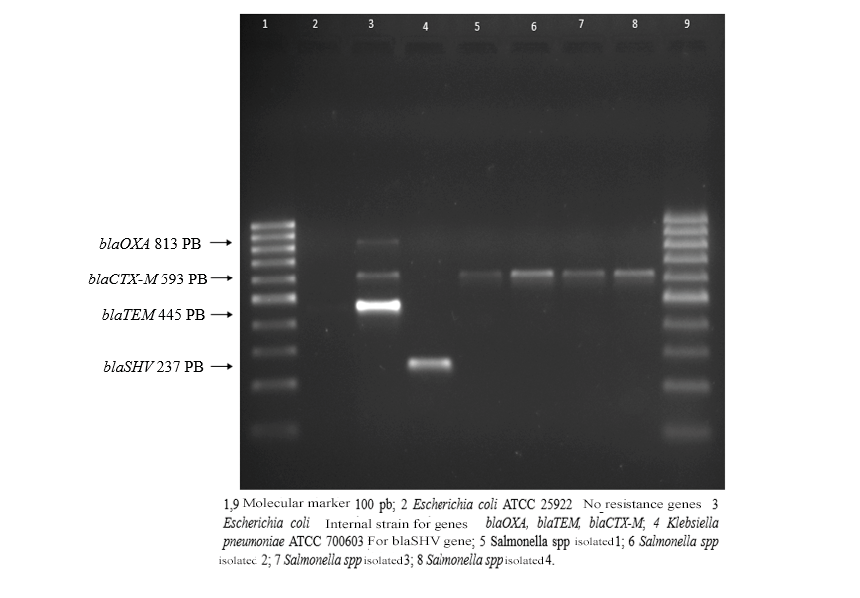
Table 1.
Resistance profile and extended-spectrum beta-lactamase (ESBL) genes.
|
Isolate |
Type of sample |
Antimicrobial resistance profile |
PCR (ESBL) genes) |
||||||||||||
|
AMP |
CRO |
CAZ |
CTX |
AMC |
STX |
MER |
IPM |
CIP |
BLEE |
OXA |
TEM |
SHV |
CTX-M |
||
|
Salmonella spp |
Super ground beef |
R |
R |
R |
R |
S |
S |
S |
S |
I |
+ |
- |
- |
- |
+ |
|
Salmonella spp |
Super ground beef |
R |
R |
R |
R |
S |
S |
S |
S |
I |
+ |
- |
- |
- |
+ |
|
Salmonella spp |
Super ground beef |
R |
R |
R |
R |
S |
S |
S |
S |
I |
+ |
- |
- |
- |
+ |
|
Salmonella spp |
Super ground beef |
R |
R |
R |
R |
S |
S |
S |
S |
I |
+ |
- |
- |
- |
+ |
Table 2.
Primer sequences for different β-lactamase resistance genes.
|
Primer names |
Primer sequence |
Size (Pb) |
Reference |
|
blaSHV |
Forward 5´- CTTTATCGGCCCTCACTCAA -3´ |
237 Pb |
(Hong Fang, 2018. DOI 10.1128/JCM.01943-07) |
|
blaSHV |
Reverse 5´- AGGTGCTCATCATGGGAAAG -3´ |
||
|
blaTEM |
Forward 5´- CGCCGCATACACTATTCTCAGAATGA -3´ |
445 Pb |
|
|
blaTEM |
Forward 5´- ACGCTCACCGGCTCCAGATTTAT -3´ |
||
|
blaCTX-M |
Forward 5´- ATGTGCAGYACCAGTAARGTKATGGC -3´ |
593 Pb |
|
|
blaCTX-M |
Reverse 5´- TGGGTRAARTARGTSACCAGAAYCAGCGG -3´ |
||
|
blaOXA |
Forward 5´- ACACAATACATATCAACTTCGC -3´ |
813 Pb |
|
|
blaOXA |
Reverse 5´- AGTGTGTTTAGAATGGTGATC -3´ |
Work Cited
Clinical And Laboratory Standards Institute. (2020). Clsi, M100 Performance Standards For Antimicrobial Susceptibility Testing. Clinical And Laboratory Standards Institute.
Divek V. T. Nair, K. V. (2018. doi:10.3390/foods7100167). Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Food.
FAO. (2020). Resistencias a los antimicrobianos en alimentos. Organización de las Naciones Unidas para la Alimentación y la Agricultura: Accesado 2022 http://www.fao.org/3/ca8275es/CA8275ES.pdf.
Hong Fang, F. A. (2018. DOI 10.1128/JCM.01943-07). Molecular Epidemiology of Extended-Spectrum B-Lactamases among Escherichia coli Isolates Collected in a Swedish Hospital and Its Associated Health Care Facilities from 2001 to 2006. Journal of Clinical Microbiology.
Margarita Terentjeva, J. A. (2017). Prevalence and antimicrobial resistance of Salmonella in meat and meat products in Latvia. Ann Agric Environ Med. 2017;24:317–321 DOI: 10.5604/12321966.1235180.
OMS. (2016). Plan De Acción Mundial Sobre La Resistencia A Los Antimicrobianos. Organizacion mundial de la salud: http://apps.who.int/iris/bitstream/10665/255204/1/9789243509761-spa.pdf
Rodríguez, J. A. (2000). Métodos básicos para el estudio de la sensibilidad a los antimicrobianos. Procedimientos en Microbiología Clínica. Recomendaciones de la Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica.
Samuel Vilchez, D. R. (2009). Prevalence of diarrhoeagenicEscherichia coliin children from Leon, Nicaragua. Journal of Medical Microbiology, 58,630–637. DOI 10.1099/jmm.0.007369-0.
Wallace H. Andrews, H. W. (2019). BAM Chapter 5: Salmonella. En Bacteriological Analytical Manual (BAM) Food and Drug Administration (FDA). Ultimo acceso 2019. Disponible en: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-Salmonella.
